Welcome to icebreaker’s documentation!¶
Icebreaker provides Python interface for the JBEI ICE sample manager.
See the full API documentation here
Installation¶
Icebreaker is written for Python 3+. You can install icebreaker via PIP:
sudo pip install icebreaker
Alternatively, you can unzip the sources in a folder and type
sudo python setup.py install
Example of use¶
In this example we assume that we are a lab who wants to find primers from its database to sequence a given construct. We will (1) pull all our primers from ICE, (2) find which primers are adapted to our sequence, using the Primavera package, and (3) we will ask ICE for the location of the selected primers.
Connexion to an ICE instance¶
You can connecct to your ICE instance using either an API token (see below for how to create a token), or an email/password authentication.
import icebreaker
# CONNECT TO ICE
configuration = dict(
root="https://my.ice.instance.org",
api_token="WMnlYlWHz+BC+7eFV=...",
api_token_client = "icebot"
)
ice = icebreaker.IceClient(configuration)
Or:
# CONNECT TO ICE
configuration = dict(
root="https://my.ice.instance.org",
email="michael.swann@genomefoundry.org",
password = "ic3ic3baby"
)
ice = icebreaker.IceClient(configuration)
The configuration can also be written in a yaml file so you can write
IceClient('config.yml') where config.yml reads as follows:
`
root: https://my.ice.instance.org
email: michael.swann@genomefoundry.org
password: ic3ic3baby
`
Extracting all records from a folder¶
Next we pull all primers in the database:
# FIND THE ID OF THE FOLDER WITH ALL PRIMERS
primers_folder = ice.get_folder_id("PRIMERS", collection="SHARED")
# GET INFOS ON ALL ENTRIES IN THE FOLDER (PRIMER NAME, ID, CREATOR...)
primers_entries = ice.get_folder_entries(primers_folder)
# GET A BIOPYTHON RECORD FOR EACH PRIMER
primers_records = {primer["id"]: ice.get_record(primer["id"])
for primer in primers_entries}
Primer selection with Primavera¶
Next provide this information to Primavera and select some primers (see the Primavera docs):
from primavera import PrimerSelector, Primer, load_record
available_primers = [
Primer(sequence=primers_records[entry['id']].seq.tostring(),
name=entry['name'],
metadata=dict(ice_id=entry['id']))
for entry in primers_entries
]
constructs = [load_record("RTM3_39.gb", linear=False)]
selector = PrimerSelector(read_range=(150, 1000), tm_range=(55, 70),
size_range=(16, 25), coverage_resolution=10,
primer_reuse_bonus=200)
selected_primers = selector.select_primers(constructs, available_primers)
Finding available samples¶
Finally we look for available samples for each primer:
selected_primers = set(sum(selected_primers, []))
for primer in selected_primers:
ice_id = primer.metadata.get("ice_id", None)
if ice_id is not None:
samples = ice.get_samples(ice_id)
if len(samples) > 0:
location = icebreaker.sample_location_string(samples[0])
print("Primer %s is in %s." % (primer.name, location))
Result:
Primer PRV_EMMA_IN00042 is in PRIMER_PLATE_1/E06.
Primer PRV_EMMA_IN00043 is in PRIMER_PLATE_1/F06.
Primer PRV_EMMA_IN00028 is in PRIMER_PLATE_1/G04.
Primer PRV_EMMA_IN00060 is in PRIMER_PLATE_1/G08.
Primer PRV_EMMA_IN00064 is in PRIMER_PLATE_1/C09.
Primer PRV_EMMA_IN00038 is in PRIMER_PLATE_1/A06.
Primer PRV_EMMA_IN00068 is in PRIMER_PLATE_1/G09.
API¶
-
class
icebreaker.IceClient(config, cache=None, logger='bar')[source]¶ Session to easily interact with an ICE instance.
Methods
-
set_api_token(client, token)[source]¶ Set a new API token (and erase any previous token / session ID)
Examples
>>> ice_client.set_api_token('icebot', 'werouh4E4boubSFSDF=')
-
get_new_session_id(email, password)[source]¶ Authenticate and receive a new session ID.
This is automatically called in IceClient if the config contains an email and password.
-
request(method, endpoint, params=None, data=None, files=None, response_type='json')[source]¶ Make a request to the ICE server.
This is a generic method used by all the subsequent methods, and wraps the
requests.requestmethodParameters: method
One of “GET”, “POST”, etc. see documentation of the Request library.
endpoint
Part of the address after “http/…/rest/”.
params
Parameters to add to the url, as a dict (the ones after “?=”)
data
Json data to add to the request
files
response_type
Use “json” if you expect JSON to be returned, or “file” if you are expecting a file.
Examples
>>> ice = IceClient(...) >>> ice.request("GET", "GET", "folders/12/entries", params=dict(limit=20))
-
create_part_sample(part_id, plate_name, well, depositor='auto', sample_label='auto', tube_display='auto', sample_barcode='auto', plate_type='PLATE96', assert_sample_created=True)[source]¶ Create a new sample for a part.
This will fail silently if a sample already exists in the given well.
Returns: query_result
A dict of the form
{resultCount: 0, data:[{location:}, ...]}where thedatalist seems to contain the same as the result ofself.get_part_samples. the last item in the list will be the freshly added sample.
-
delete_part_sample(part_id, sample_id)[source]¶ Delete a given sample for a given part.
Note that if this is the last sample from a plate, the plate will disappear (stop showing in self.get_plates_list)
-
search(query, limit=None, batch_size=50, as_iterator=False, min_score=0, entry_types=(), sort_field='RELEVANCE')[source]¶ Return an iterator or list over text search results.
Parameters: query
Text to query
limit
limit on the number of entries to fetch
batch_size
How many entries to get at the same time at each connexion with ICE ideal may be 50
as_iterator
If True an iterator is returned, if False a list is returned.
min_score
Minimal score accepted. The search will be stopped at the first occurence of a score below that limit if sort_field is “RELEVANCE”.
Returns: entries_iterator
An iterator over the successive entries found by the search.
-
find_entry_by_name(name, limit=10, min_score=0, entry_types=('PART', 'PLASMID'))[source]¶ Find an entry (id and other infos) via a name search.
Note that because of possible weirdness in ICE searches, it is not guaranteed to work.
Parameters: name
Name of the entry to retrieve
limit
Limitation on the number of entries to consider in the search. Ideally, get just 1 result (top result) would be sufficient, but you never know, some parts with slightly different names could rank higher.
entry_types
List of acceptable entry types. The less there is, the faster the search.
Returns: entry_info, None
In case of success.
entry_infois a dict with part ID, owner, etc.- None, (“Multiple matches”, [entries_ids…])
In case several ICE entries have that exact name
- None, (“No match”, [“suggestion1”, “suggestion2” …])
Where the suggestions are entry names in ICE very similar to the required name.
-
get_folder_entries(folder_id, must_contain=None, as_iterator=False, limit=None, batch_size=15)[source]¶ Return a list or iterator of all entries in a given ICE folder.
Each entry is represented by a dictionnary giving its name, creation date, and other infos.
Parameters: folder_id
ID of the folder to browse.
must_contain
If provided, either the id or the name of the entry (part) must contain that string.
limit
If provided, only the nth first entries are considered.
batch_size
How many parts should be pulled from ICE at the same time.
as_iterator
If true, an iterator is returned instead of a list (useful for folders with many parts)
-
get_collection_entries(collection, must_contain=None, as_iterator=False, limit=None, batch_size=15)[source]¶ Return all entries in a given collection
-
get_collection_folders(collection)[source]¶ Return a list of folders in a given collection.
Collection must be one of: FEATURED PERSONAL SHARED DRAFTS PENDING DELETED
-
change_user_password(new_password, user_id='session_user')[source]¶ Change the password of a user (current session user by default)
-
restrict_part_to_user(part_id, user_id='current_user')[source]¶ Remove all permissions that are not from the given user.
-
create_part(name, description='A part.', pi='unknown', parameters=(), **attributes)[source]¶ Create a new part.
Parameters: name
Name of the new part
-
create_folder_permission(folder_id, group_id=None, user_id=None, can_write=False)[source]¶ Add a new permission for the given folder.
Parameters: folder_id
ID of the folder getting the permission
user_id, group_id
Provide either one to identify who gets the permission
can_write
Allows users to both add files to the folder and overwrite data from files in the folder (!! use with care).
-
delete_folder_permission(folder_id, permission_id)[source]¶ Remove a permission attached to a given folder.
-
add_to_folder(entries_ids=(), folders=(), folders_ids=(), remote_entries=())[source]¶ Add a list of entries to a list of folders.
Parameters: entries_ids
List of entry IDS
folders
List of full folder infos. (NOT folder ids). I guess this is to allow folders on remote ICE instances. Confusing, but you can use
folders_idsinstead.folder_ids
List of folder IDs that can be provided instead of the
foldersinfos list.
-
attach_record_to_part(ice_record_id=None, ice_part_id=None, record=None, record_text=None, filename='auto', record_format='genbank')[source]¶ Attach a BioPython record or raw text record to a part
Parameters: ice_record_id
A uuid (like aw3de45-sfjn389-lng563d…) that identifies the record attachment of the part in ICE. It is generally the field “recordId” when you retrieve a part’s infos in ICE. If you have no clue, leave this to None and provide a
ice_part_idinstead.ice_part_id
The id of an ICE entry that can be provided instead of the
ice_record_id.record
A Biopython record
record_text
Raw text from a FASTA/Genbank record.
filename
If set to “auto”, will be “record_id.gb” for genbank format.
record_format
When providing a fasta format in record_text, set this to “fasta”.
-
get_user_groups(user_id='session_id')[source]¶ List all groups a user (this user by default) is part of.
-
find_parts_by_custom_field_value(parameter, value)[source]¶ Find all parts whose (extra) field “parameter” is set to “value”
-
rebuild_search_indexes()[source]¶ Rebuild the search indexes.
An OK response does not mean that it is finished, just that the rebuild was scheduled.
-
get_part_custom_fields_list(part_id)[source]¶ Return a list of all custom fields for a given part.
Returns a list of the form
[{name: 'field1', value: 321}, ...]
-
get_part_custom_field(part_id, field_name)[source]¶ Return the value for a part’s custom field.
The value will be a list if the part has several values attached to that field name.
-
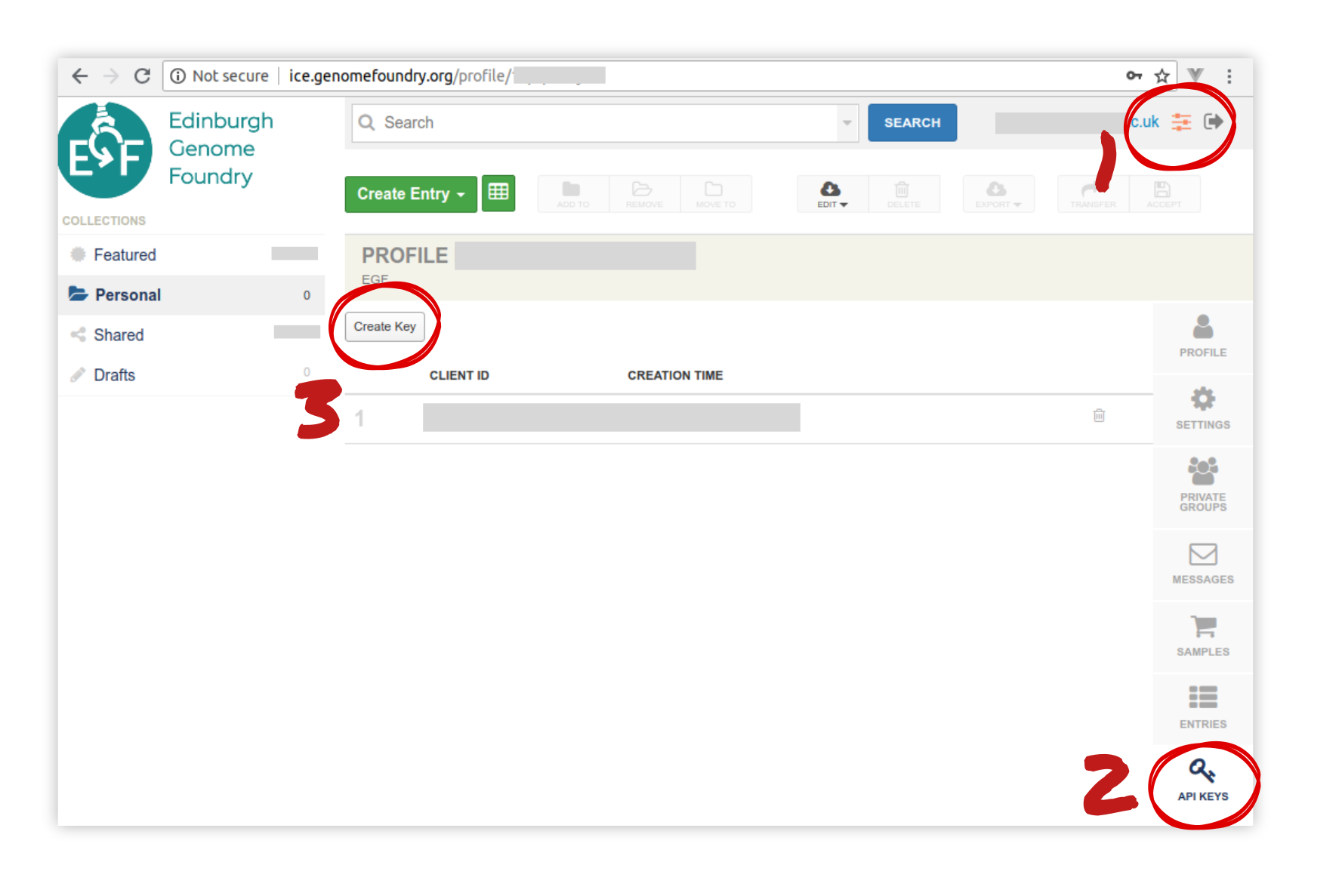
Getting an ICE token¶
There are several ways to get ICE tokens. We suggest you create one throug the web interface as follows (see screenshot for indications):
- Create an account with administrator rights
- Go to the administrator panel
- Click on “API keys”
- Click on “create new”. Note everything down !
License = MIT¶
Icebreaker is an open-source software originally written at the Edinburgh Genome Foundry by Zulko and released on Github under the MIT licence (¢ Edinburg Genome Foundry). Everyone is welcome to contribute !
More biology software¶

Icebreaker is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.